Protocols | Overview
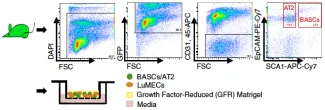
Isolation and sorting for bronchioalveolar stem cells (BASCs)
BEFORE BEGINNING, you will need:
- Avertin (1X stock in 4°C under bench)
- Forceps, surgical scissors, pins, dissecting platform (Styrofoam lid wrapped in foil, covered with a paper towel)
- Cold PBS
- 10, 3, and 1 mL syringes
- 20G needles
- 10% Formalin (RT)
- Dispase (aliquots in -20°C; **THAW IN 50°C WB, remove immediately to ice once thawed**)
- 1% LMP Agarose (RT, **THAW IN 50°C WB, leave in bath until ready to use)
- Collagenase/dispase (-20°C below bench)
- DNAse (-20°C below bench; 1% solution; check unit activity of each lot number)
- PF10 (make 50 mL by adding 5 mL FBS (4°C below bench) to 45 mL PBS)
- 100 and 40 μm filters (above bench)
- Bucket with ice
- 50 mL conical tubes
Steps
- anesthetize mouse with avertin, test with forceps (toe pinch) to make sure under enough
- spray down mouse with 70% ethanol
- quickly cut into ribcage, perfuse 10 mL of PBS (ice cold) through right ventricle until lungs cleared of blood (cut a slit in left ventricle to allow blood to leave)
- cut out heart to euthanize mouse
- expose trachea, place forceps under trachea to keep exposed
- inject dispase (BD, undiluted liquid, aliquots in -20°C) into trachea (go between the cartilage rings) just until the lungs inflate — you should be able to do this with less than 2 mL
- follow with tracheal injection of 0.5-1 mL of 1% LMP agarose (made in H2O)
- place a few pieces of ice on lungs, leave everything there for a few minutes as you prep needles, etc. for the next mouse
- dissect out the lungs by gently tugging on the trachea while snipping away the connective tissue; leave lungs intact and place in Petri dish on ice with PBS
- trim other tissues off lung, dissect off each lobe, and mince lung tissue into small pieces inside of a 50 mL conical tube. For each lung sample, add 3 mL PBS.
- add 60uL collagenase/dispase for every 3mL of PBS above (from Roche, 100 mg/mL in H2O stock stored in -20°C below bench; powder in cold room) to PBS; rotate 45 min @ 37°C; final collagenase/dispase concentration is ~2 mg/mL
- Place digested tissue on ice. Add 7.5 uL DNAse per 3 mL of digest solution (from 1% stock stored in -20°C, which is 10 mg/mL); final DNAse concentration is ~0.025 mg/mL
- Invert several times to mix. Leave on ice to cool for no more than 5 minutes.
- Into a 50 mL tube, filter digested tissue through a 100 um filter, then a 40 um filter
- Spin 6 min @ 800 rpm in Beckman centrifuge
- Discard supernatant; resuspend pellet in 2 mL PF10 (10% FBS in PBS) and count/stain cells OR proceed to RBC lysis protocol if you need to eliminate RBCs
Red blood cell lysis
- Resuspend cell pellet from above in 1 mL of lysing solution (0.15 M NH4Cl, 10mM KHCO3, 0.1 mM EDTA, in 1L distilled H2O; filtered with 0.45 um filter and stored at RT)
- Lyse 90 sec at RT
- Add 6 mL of DMEM without serum (refrigerator below bench); mix
- Add 0.5 mL FBS slowly to bottom of tube by inserting pipet tip all the way through the resuspended cell solution
- Centrifuge undisturbed layers for 6 min @ 800 rpm
- Aspirate the supernatant; resuspend in PF10 for counting or staining
Surface staining
- Stain 1 million cells in 100 ul PF10 (~100uL to 300uL per mouse) for 15 minutes with appropriate amount of antibody on ice; touch spin in microcentrifuge for ~10 sec, aspirate supernatant, wash with ~3x volume of stain solution PF10, touch spin, aspirate sup, resuspend in 300 uL PF10 for FACS
- Repeat for secondary antibody if necessary
- Use single-stained controls for every antibody you use and a negative control (no staining) for every mouse/genotype/situation for which you will sort cells. This is for compensation. Cells for these should come from a control sample; dilute the original sample such that after 100 uL aliquots are placed into each control tube, 500 uL of the sample remain for staining.
- Stain with Sca-1 (usually FITC conjugated), CD45 Biotin (note it will depend on the strain of mouse which CD45 to use; for 129 and B6 strains, use CD45.2), Pecam Biotin, SA-PE (secondary), then 7AAD (see protocol below) to exclude dead cells
- Ab dilutions:
- 1° Sca-1-FITC à 1:100
- 1° CD31 (Pecam)-Biotin à 1:100 (i.e. 5 uL in 500 uL)
- 1° CD45-Biotin à 1:100
- 2° SA-APC à 1:200 (i.e. 2.5 uL in 500 uL)
- better to use SA-APC instead of PE for better compensation (PE and FITC are more difficult to compensate than APC and FITC)
7AAD staining
- Resuspend the cells in appropriate amount of PF10. Add 7AAD so that it is 1:300 dilution. (7AAD comes as a powder from Molecular Probes in Oregon, A-1310, resuspend in 500 uL DMSO (this makes a 2 mg/mL stock), store in freezer and thaw before each use)
- Incubate cells on ice for 15 minutes
- Touch spin, wash as before
- Resuspend cells in PF10 for sorting
- Filter the cell/PF10 suspension through the blue (40u) filters, either directly into FACS tubes or into 50mL conical and then transfer to appropriate tube for FACS. (depends on the density of your cells)(be sure to use the #2063 tubes for Moflos, doesn’t matter for Aria). This is very important so that you won’t clog the FACS machine!
- *if you are just looking for GFP+ cells, you don’t have to do any staining — just count and put approximately 1 million cells per 300 uL PI solution or PF10 solution for FACS
- *keep cells on ice as much as possible; bring tubes with media/solution you want to collect sorted cells into to the FACS facility
Cell Population Isolation
- For BASCs (Bronchio-alveolar stem cells), sort the Sca-1+ CD45- Pecam- population; this population also contains ciliated cells
- For clonal assays (use of single cells), use Sca-1+ CD45- Pecam- and CD34+ cells (just add CD34 staining into the mix above) — this will enrich for your ability to isolate single cells capable of giving rise to colonies.
- For AT2 cells, sort the Sca-1- CD45- Pecam- population that has high autofluorescence in FITC channel (in your negative control, you should be able to see 2 populations falling out — highly autofluorescent (still live) and low autofluorescent
Sorting cells
- For cultures, you can sort directly onto a plate or into a tube and then plate by hand
- For sorting into tubes, prepare collection tubes ahead of time: 3% BSA in PBS, made fresh or filtered, fill up the collection tube and let stand at least one hour (or overnight in 4°C). Aspirate this solution, add about 200 uL of media into which to sort the cells.
- ***It seems very important to reduce the pressure setting on the FACS machine for BASCs to grow. We have been using a Moflo sorter, 30Psi with success. Other machines/pressure settings have not yet been verified. When you make the FACS appointment, you must request the pressure setting to be lower, they cannot just change it at your appointment.
Cytospin
Sorted or cultured cells can be stained after cytospin at 600 rpm 3 min, fixation and permeabolization with CytoFix/CytoPerm (Pharmingen) for 20 min at 4°C (if on matrigel, fix at RT). Followed by washing in PBS/0.2% Tx-100, 3 x 5 min.; blocking and primary antibody. (**See immunofluroescence protocol)
Epithelial cultures
Cells can be plated in DME+/HEPES/10% FBS/Pen-Strep/L-glutamine on 96-well plates coated with 100 mL Matrigel (for differentiation)(Becton Dickinson) or irradiated DR4 MEFs (for maintenance of undifferentiated state, colony formation assays, etc.).
Make the feeder plate the day before you sort cells onto it.
Feeding BASC cultures: start with a plate of feeders and 100uL media, sort cells on top (Day 0)
- 4 days later, supplement the plate by adding 100uL additional media; do not aspirate the media off — it may disturb the cells that are trying to adhere and form colonies and disturbs the feeder layer
- cells can be infected at 4-5 days after plating for lentiviral or retroviral work-spinfection has worked well
- do not aspirate the media off until 7-10 days after the cells have been plated
For Matrigel, best to plate at least 10,000 cells per 96-well; for feeders, plate 1000 cells per 96-well unless for limiting dilution. Coat the 96-well with 100 uL Matrigel that has been thawed on ice, place plate in incubator for 20 minutes (don’t do more or less time, important for the Matrigel to solidify correctly), then pipette the cell mixture on top of the Matrigel. Cells do not do well on larger well formats, perhaps unless same cell density is maintained.
Adeno infection
- Matrigel-plated cells have typically been infected on the day after plating: aspirate media, add media with adeno, fresh media next day;
- for feeder plates, best to infect the day before you are going to replate. Make a concentrated stock of the AdGFP and supplement the existing wells (for example, if about 150uL on cells at the time, make a 4X virus mix and add 50uL per well into the wells with media — this way you don’t have to aspirate)
- when replating cells for secondary, tertiary colonies, etc.:
- do not spin the cells — just wash with PBS, trypsinize (10 minutes) with 50uL, add 100uL media, mix well and pipette through 40u filter, take directly to FACS
- do not stain for PI or 7AAD; 7AAD does not work on the cultured cells, and we don’t know yet if PI affects the viability of the cells
- do not replate based on AdGFP signal, even if you used it to score colonies; feeders are still infected, and may be just as bright as your cells of interest; we also don’t know yet if bringing along any non-epithelial cells is needed for secondary colonies, etc.
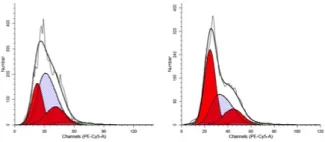
Cell Cycle flow cytometry
7AAD Cell Cycle Flow Cytometry
- Aspirate media
- Add 5mL 1x PBS and swirl plate to wash cells
- Aspirate PBS
- Add 1mL 0.25% Trypsin
- Incubate plate for 4 minutes at room temperature
- Shake plate to loosen cells from surface of plate
- Add 4mL fresh media (RPMI, 10% serum)
- By tilting plate, pipet cells into a puddle at one edge
- Move all 5mL of cells to a 15cc conical tube
- Place tube on ice
- For etoposide, leave all 5 mL, for all others, remove 4 mL (save 1mL per sample)
- Spin down 5 min 1000 rpm
- Resuspend in 300 µL PF10
- Add dropwise to 700 µL ICE COLD 70% EtOH in eppendorf, vortex slowly while adding
- Let sit at 4˚C for at least 1 hour, 24-48 hours is best
- Pellet cells by pulse spin or 2 min 6000rpm
- Resuspend in RNAse A, 250uL/tube of 1mg/mL RNAse A diluted in PBS
- Add up to 1mL PBS and shake to wash
- Pellet cells by pulse spin or 2 min 6000rpm
- Resuspend in 250µL/tube of 2µg/mL PI or 7AAD solution (10µL 7AAD stock per 1mL buffer)
- Flow as PE-Cy5 for 7AAD, or PE for PI (though some BD flow cytometers have a PI specific setting) (linear)
- Analyze with ModFit (flow core) LT if you can get the software, it’s the best!
